Help beat coronavirus – plug your computer’s performance into the joint computation
2 min read
The nearly 20-year-old Folding @ HOme project, backed by scientists at Stanford University, has focused on simulating protein folding, drug testing, and other molecular physics from the outset. Everything is solved by means of distributed calculations, which involve both powerful supercomputers and common computers of users from all over the world.
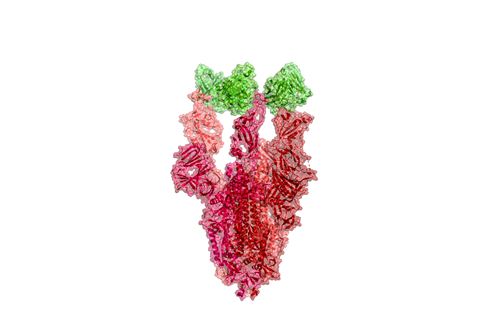
Now Folding @ Home has announced the launch of the program for fighting against coronavirus Covid-19 (2019-NCOV). It will analyze the structure of the virus and above all the spike protein. It results in the virus being able to attach itself quickly and easily to protein receptors in the lung cells.
The aim is to develop a type of protein commonly referred to as a “therapeutic antibody” that can precisely block the virus protein against attachment to receptors and thus prevent lung cell infection itself.
But proteins are a dynamic and complex structure, so a lot of combinations need to be simulated to reveal how it actually works. Since there is already a model for older low-resolution SARS-CoV, research can be expected to go faster. There are many similarities between the two viruses.
The higher the computing power available for scientists, the faster and more accurate simulations they can perform. You can join the project by downloading the client from the site, which is for all major operating systems and can use both the power of the processor and graphics cards. You can control how much of your computer’s performance you want to share, so you don’t have to worry about making the most of your simulation.